 At FHL we try hard to both honor our past by maintaining our core strengths and adapt to the needs of the FHL community. An example is the summer teaching program. During our strategic planning sessions two years ago we heard from many stakeholders about the need for shorter, non-credit-bearing, skill-building workshops. Such workshops could especially benefit graduate students, who in today’s climate have a harder time finding the time and funds for 5-week “basic education” classes, instead needing to focus on their own research or on training that will promote that research. In response, this summer we offered three 3-week workshops on diverse topics. Below, Ryan Kelly reports on one of those workshops, which was very successful based on many measures!
At FHL we try hard to both honor our past by maintaining our core strengths and adapt to the needs of the FHL community. An example is the summer teaching program. During our strategic planning sessions two years ago we heard from many stakeholders about the need for shorter, non-credit-bearing, skill-building workshops. Such workshops could especially benefit graduate students, who in today’s climate have a harder time finding the time and funds for 5-week “basic education” classes, instead needing to focus on their own research or on training that will promote that research. In response, this summer we offered three 3-week workshops on diverse topics. Below, Ryan Kelly reports on one of those workshops, which was very successful based on many measures!
Best,
Dr. Megan Dethier, FHL Director
Make your gift today
The Molecular Memory of Water
by Ryan Kelly
Trained as both an ecologist and a lawyer, Ryan Kelly has a broad set of interests, focused both on hard scientific data and policymakers’ use of those data. Ryan joins genetic and ecological research with real-world implementation in law and policy, particularly with respect to environmental monitoring and resource management. He is the Director of the eDNA Collaborative, and Associate Director of the School of Marine and Environmental Affairs within the University of Washington’s College of the Environment.
Every living thing leaves behind traces of its existence in the form of genetic material – DNA or RNA – in its environment. Scientists have long known that it is possible to sequence DNA out of a sample of water or soil – indeed, this is something microbiologists have done for decades. But until the past decade or so, it was far less clear that we could learn a lot about the ecology of larger-bodied species using the same techniques. Just as is the case for bacteria and other single-celled organisms, every sea star, worm, fish, or alga is represented in the genetic library of a water sample taken nearby. We call this genetic material “environmental DNA,” eDNA for short.
We are all constantly surrounded by this high-resolution map of the living world, and scientists are only just beginning to figure out how to make use of it to answer basic ecological and management questions. For example, eDNA recently featured in a NOAA stock assessment for the first time, helping report the distribution and abundance of hake (Merluccius productus) along the west coast of the US and Canada (Johnson et al. 2025). And within Puget Sound, eDNA has proved an effective tool for monitoring everything from invasive green crab (Carcinus maenas, Keller et al. 2022) to harmful algal blooms (Jacobs-Palmer et al. 2021).

With a grant from the Packard Foundation, we started the eDNA Collaborative at UW in 2022 to help move these kinds of eDNA techniques out of the lab and into routine practice around the country and around the world. Pretty much immediately, we realized that if we wanted to make eDNA generally useful, we would need to help lower barriers that include 1) significant molecular expertise and training, and 2) expensive laboratory reagents and equipment. We’ve tried to tackle these through microgrants to individual researchers – to date, more than 150 up-and-coming scientists in 42 countries have received equipment or training – but for 2025 we wanted to focus on a single, hands-on experience, and of course, we wanted to do it at FHL.
In July, we welcomed 14 workshop participants from 11 countries, traveling from points as far afield as Patagonia, the Galapagos, Benin, Côte d’Ivoire, Indonesia, Hong Kong, and Brazil (Figure 1). They represented a mix of career stages — full professors to new grad students — and came to work together with our amazing six-person UW teaching team to move the field of eDNA research forward. Our teaching staff included Eily Allan, Aden Ip, Pedro Brendão, Gled Guri, Shana Hirsch, and me, with some help from Kate Bertko, and was supported both by the Packard Foundation and Oceankind.
We could have held an eDNA class anywhere, given that the planet is covered in genetic information awaiting study. In that way, the workshop was different than, say, the FHL Invertebrate Zoology course, which is closely tied to the region’s particular fauna. But making ecological meaning out of a string of Latin names (after matching their DNA sequences to a database) requires developing some familiarity with the organisms present – that is, developing the sense of place that is important to us all as biologists. FHL offered the chance to do just this, with a fit-for-purpose genomics teaching lab in sight of our sampling site, the FHL dock. Our experimental design was simple: testing for wholesale community differences in the day vs. the night, as well as looking for an effect of the favorite FHL nighttime activity, nightlighting, in which enthusiasts hang an LED light in the water off of the FHL dock to see the reactions of invertebrate larvae, juvenile fishes, and other nektonic marine life.
Teaching a three-week workshop (rather than a full-length course for UW credit) worked especially well for us because of our participant population: we aimed at folks a bit further along in their careers, many of them with jobs and families (Figure 2). They simply couldn’t leave their real lives for five weeks. The shorter, high intensity format let us attract professionals that are at the center of their own networks at home, which will amplify the workshop’s impact as its lessons ripple outward in the months and years to come.

But this short format meant we needed to move fast in order to have meaningful data in hand during the course. One way that eDNA work can be difficult is the lag times common between when samples are collected and when usable data is available. In some contexts where lab facilities are in short supply, this lag can be months or years. Here we had an opportunity to hone our quick-turnaround skills, doing the sequencing immediately using small sequencing machines made by Oxford Nanopore. This technology has been around for years, but the machines can be prone to relatively high error rates, the relevant software can be difficult to use, and the downstream data can be difficult to handle. To head off these problems, staff scientist Aden Ip led the charge to code a full-scale bioinformatics pipeline that would allow the class to process data in near-real time.
We began immediately upon arriving at FHL, sampling eDNA out of seawater for a week and sequencing it on site to provide data on more than 700 species from across the tree of life. In addition to the diatoms, kelp, fish, and polychaetes one might expect, we also picked up gray whale, northern fur seal, and — particularly exciting for many of us — the critically endangered sunflower star, Pycnopodia helianthoides. We were able to see fairly clear day vs night differences in the communities of invertebrates, algae, and fishes, and in the bargain, we got a very good idea of how stable the eDNA signals are over time in the nearshore marine community at FHL (Figure 3).
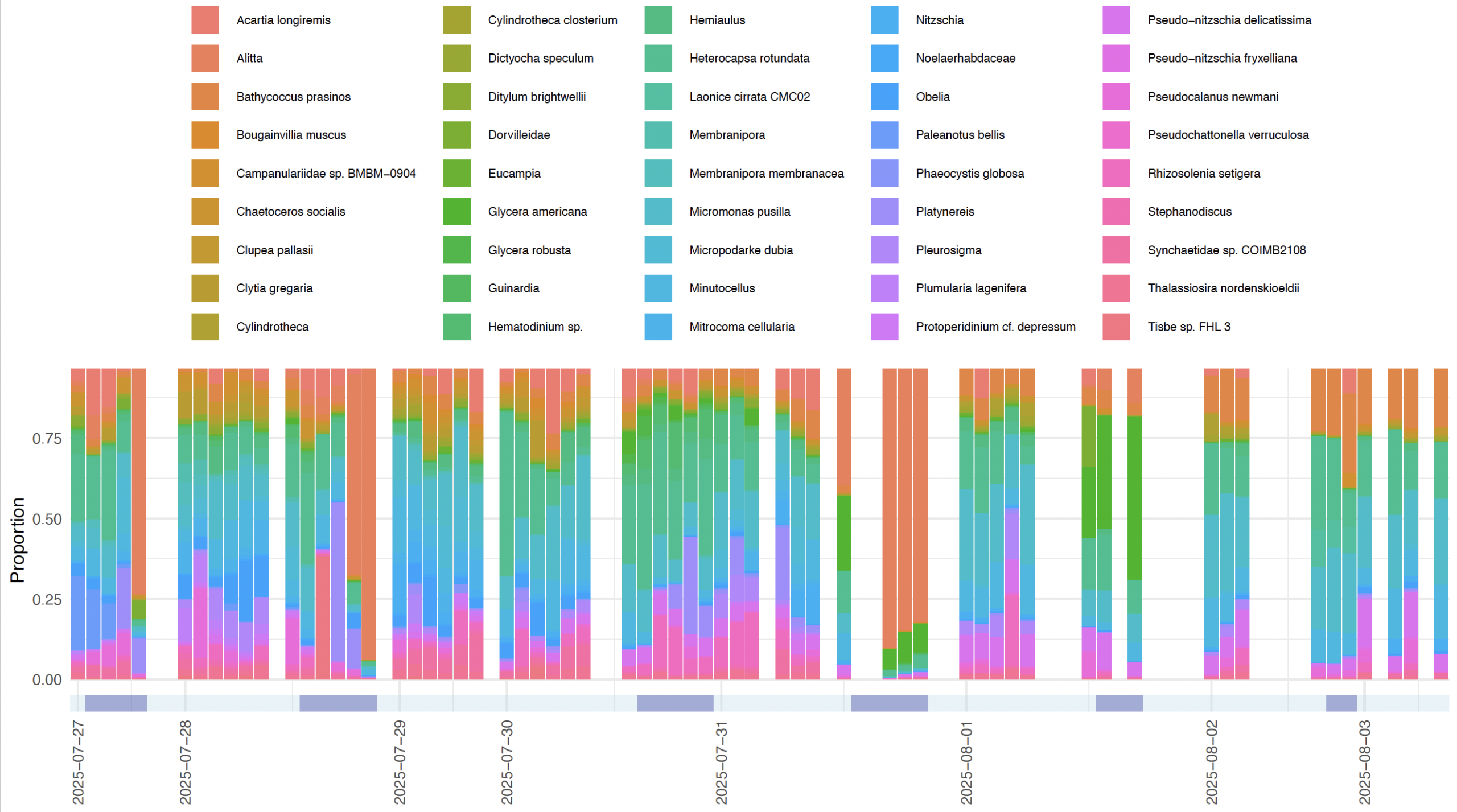
To have gone from being total strangers in late July to carrying out a full-scale experimental eDNA project by mid-August – from field sampling through lab work, sequencing, bioinformatics, and multivariate community analysis – was a pretty amazing experience for all of us. The group has kept in touch via email and Slack and is working toward a peer-reviewed publication, led by one of our workshop participants, Kingsly Chuo Beng. Already, some of us are planning our next collaborations, which is especially exciting given the eDNA Collaborative’s goal of developing networks of practice, and I know we all look forward to working to advance eDNA techniques beyond what is currently possible.
References:
Jacobs-Palmer E., Gallego R., Cribari K., Keller A.G., and R.P. Kelly. 2021. Environmental DNA Metabarcoding for Simultaneous Monitoring and Ecological Assessment of Many Harmful Algae. Frontiers in Ecology and Evolution: 9-2021. https://doi.org/10.3389/fevo.2021.612107
Johnson K.F., Edwards A.M., Berger A.M., Grandin C.J., and C.R. Wetzel. 2025. Status of the Pacific Hake (whiting) stock in U.S. and Canadian waters in 2025. Prepared by the Joint Technical Committee of the U.S. and Canada Pacific Hake/Whiting Agreement, National Marine Fisheries Service and Fisheries and Oceans Canada. 286 p.
Keller A.G., Grason E.W., McDonald P.S., Ramón-Laca A., and R.P. Kelly. 2022. Tracking an Invasion Front with Environmental DNA. Ecological Applications 32(4): e2561. https://doi.org/10.1002/eap.2561
Tide Bites is a monthly email with the latest news and stories about Friday Harbor Labs. Want more? Subscribe to Tide Bites or browse the archives.